21 Appendix
This is a parking place for content that should be incorporated into other parts of the book
21.1 Healthy Microbiome
2024-10 from Gut Microbiome for Health
Another new study by Liping Zhao and colleagues at the Rutger Center for Microbiome Analysis, along with international collaborators, has utilized artificial intelligence models for identifying a set of gut microorganisms that play a critical role in digestion, immune responses, and mental health. What is new with the analysis is that it is based on genome-specific analysis and database independence, and it is focused on stable gut microbiome interactions4.
The core microbiome’s structure includes two distinct groups of bacteria (i.e., the Foundation Guild and the Pathobiont Guild) that compete with each other as an indicator of health and help differentiate cases from controls across 15 diseases across three continents and predict immunotherapy outcomes. Stable interactions within gut microbiome members appear more relevant than the abundance of microorganisms. These findings open a new potential way to disease prediction and classification and manage microbiome-related diseases through specific interventions that target the core microbiome related to health4.
21.2 Microbes in the air
An international team of researchers found hundreds of different genera of fungi and bacteria at up to 3,000 meters, including many human pathogens: Escherichia coli, Serratia marcescens, Prevotella melaninogenica, Staphylococcus epidermidis, Staphylococcus haemolyticus, Staphylococcus saprophyticus, Cutibacterium acnes, Clostridium difficile, Clostridium botulinum, Stenotrophomonas maltophilia, Shigella sonnei, Haemophillus parainfluenzae and Acinetobacter baumannii and health-relevant fungi such as Malassezia restricta, Malassezia globosa, Candida parapsilosis and Candida zeylanoides, Sarocladium kiliense, Cladosporium halotolerans, and Cladosporium herbarum.
https://www.pnas.org/doi/10.1073/pnas.2404191121
21.3 Diet Effectiveness Depends on Microbes
Stanford’s Michael Snyder and his lab found Distinct factors associated with short-term and long-term weight loss induced by low-fat or low-carbohydrate diet intervention
Interestingly, we observe minimal dietary differences between those who succeeded in long-term weight loss and those who did not. Instead, proteomic and gut microbiota signatures significantly differ between these two groups at baseline.
21.4 Database of Food Microbes
Unexplored microbial diversity from 2,500 food metagenomes and links with the human microbiome Carlino, NiccolòAlvarez-Ordóñez, Avelino et al. Cell, Volume 0, Issue 0
From Cell August 2024:
Here, we present curatedFoodMetagenomicData (cFMD), an open-access resource that collects food-associated microbial data to support the use of metagenomics in food science. The current release comprises 2,533 food metagenomes with standardized metadata, 1,950 of them newly sequenced within the MASTER EU Consortium. We generated 10,112 prokaryotic and 787 eukaryotic metagenome-assembled genomes (MAGs) from food that were grouped into 1,036 prokaryotic and 108 eukaryotic species clusters, 320 of which resulted to be uncharacterized when compared with >1 M existing genomes. We included these MAGs into our pipelines for sensitive taxonomic profiling and applied it to 19,833 human metagenomes, revealing species- and strain-level overlaps along the food-human axis.
21.5 Microbes in Your Bathroom
Northwestern University scientists poured through the microbes in a typical bathroom showerhead and toothbrush to find these places harbor numerous unidentified organisms, with surprisingly little overlap among the different environments.
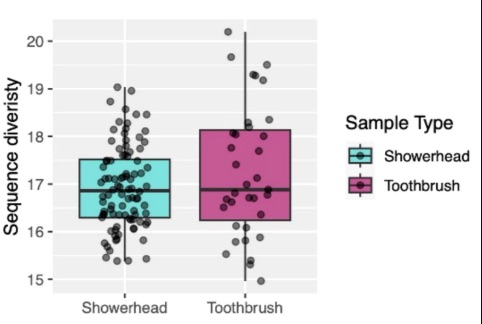
Front. Microbiomes, 08 October 2024 Sec. Environmental Microbiomes Volume 3 - 2024 | https://doi.org/10.3389/frmbi.2024.1396560
Phage communities in household-related biofilms correlate with bacterial hosts
21.6 Microbes in Your Microwave Oven
Your microwave oven has its own microbiome
Alba Iglesias, a microbiologist at the University of Valencia in Spain, and her colleagues swabbed 30 microwave ovens A total of 101 bacterial strains grew in the cultures. The dominant ones belonged to the Bacillus, Micrococcus and Staphylococcus genera, which commonly live on human skin and surfaces that people frequently touch. Human-skin bacteria were present in all three types of microwave oven, but were more abundant in the household and shared-use appliances. A few bacteria types associated with food-borne illnesses, including Klebsiella and Brevundimonas, also grew in some of the cultures from household microwaves.
Iglesias, A., Martínez, L., Torrent, D. & Porcar, M. Front. Microbiol. https://www.frontiersin.org/journals/microbiology/articles/10.3389/fmicb.2024.1395751/full (2024).
21.7 Microbes Help Animals Detect Magnetic Fields
University of Central Florida biologist Robert Fitak created a refined database of magnetic bacteria speculating that it’s microbes that help turtles and other animals navigate.
Animals’ Magnetic ‘Sixth’ Sense May Come from Bacteria, New Paper Suggests
Fitak found, for the first time, that magnetotactic bacteria are associated with many animals, including a penguin species, loggerhead sea turtles, bats and Atlantic right whales. For instance, Candidatus Magnetobacterium bavaricum regularly occurred in penguins and loggerhead sea turtles, while Magnetospirillum and Magnetococcus regularly occurred in the mammal species brown bats and Atlantic right whales.
They speculate that the microbes might live somewhere in the nervous tissue of these animals.
21.8 Most Microbes are Dormant
An estimated 60% of all microbes are lying dormant at any given moment, ready to be switched back to life when conditions are right.1
A natural protein, Balon, latches onto ribosomes to lock it in place and shut down the cell. Balon is found in around 20% of all microbes, and even more microbes might harbor genes that behave similarly.
21.9 Microbes Spread Through Dust Storms
In Unseen travelers: Dust storms may spread bacteria and fungi around the world
Dr. Shankar Chellam, professor in the Zachry Department of Civil and Environmental Engineering and A.P. and Florence Wiley Professor III at Texas A&M University, and his now former student Dr. Sourav Das have furthered previous research to identify microorganisms that might have hitched a ride in the dust plumes across the Atlantic. Dr. Daniel Spalink, assistant professor and director of S.M. Tracy Herbarium at Texas A&M, helped analyze the biology and identify bacteria and fungi in the samples.
21.10 Human Milk as a Treatment for Gut Disease
Human milk oligosaccharides (HMOs), the third most abundant component of human milk are not found in other kinds of milk, yet play a vital role in the care and feeding of Bifidobacterium.
Prolacta Bioscience is one company that makes a human-equivalent HMO intended to feed gut bacteria, and perhaps, solve numerous gut problems associated with dysfunctional microbiomes.
Seattle-based Intrinsic Medicine is currently running on clinical trial to use HMO-based drugs for treatment of Parkinsons Disease.
From Stat News Mar 2024
Biomilq, for instance, synthesizes milk using mammary cells for people who have difficulty breastfeeding. And adults have occasionally tried to tap those benefits via dietary or nutritional supplements — and even by obtaining colostrum, the first milk produced by mammals within the first 48 to 72 hours after birth. Companies such as Armra are marketing colostrum, albeit from a bovine source, in powder form.
Mentions uBiome’s Melissa Agnello for processing some samples through uBiome’s lab.
Longitudinal profiling of the microbiome at four body sites reveals core stability and individualized dynamics during health and disease
Xin Zhou, Xiaotao Shen, Jethro S. Johnson, Daniel J. Spakowicz, Melissa Agnello, Wenyu Zhou, Monica Avina, Alexander Honkala, Faye Chleilat, Shirley Jingyi Chen, Kexin Cha, Shana Leopold, Chenchen Zhu, Lei Chen, Lin Lyu, Daniel Hornburg, Si Wu, Xinyue Zhang, Chao Jiang, Liuyiqi Jiang, Lihua Jiang, Ruiqi Jian, Andrew W. Brooks, Meng Wang, Kévin Contrepois, Peng Gao, Sophia Miryam Schüssler-Fiorenza Rose, Thi Dong Binh Tran, Hoan Nguyen, Alessandra Celli, Bo-Young Hong, Eddy J. Bautista, Yair Dorsett, Paula Kavathas, Yanjiao Zhou, Erica Sodergren, George M. Weinstock, Michael P. Snyder bioRxiv 2024.02.01.577565; doi: https://doi.org/10.1101/2024.02.01.57756
21.11 Behavior
21.11.2 Alcholism and Gut Microbes
Could the gut give rise to alcohol addiction? asks Nature, describing work on Alcohol Use Disorder by Sophie Leclercq, a biomedical scientist at the Catholic University of Louvain in Brussel, and others.
Leclercq thinks that 30–40% of cases of AUD might have a gut-related component that could be targeted for treatment. A key challenge is determining exactly which components to target — it is as yet unclear what constitutes a ‘good’ microbiome. Day’s analysis suggests that bacteria such as Lactobacillus, were in abundance in people with AUD, whereas Akkermansia and some others were low.
The gut bacteria Lactobacillus, for example, can produce GABA; Enterococcus can produce serotonin; and Bacillus can make dopamine. Short-chain fatty acids (SCFAs) released when dietary fibre is fermented by bacteria in the gut also have neuroactive properties.
Review article (2023): Gut Microbiota in Anxiety and Depression: Unveiling the Relationships and Management Options
A greater incidence of depression is substantially linked to a lower protein consumption than recommended. A 10% increase in protein consumption was shown to reduce the incidence of depression considerably in South Korea and in the United States. Several biological explanations have linked the intake of protein and depression. These theories are supported by the fact that tryptophan, an amino acid, is a precursor of serotonin.
21.12 Beyond Microbes
‘Obelisks’: Entirely New Class of Life Has Been Found in The Human Digestive System Stanford University biologist Ivan Zheludev searched millions of published genome and identified at least 30K different Obelisks that appeared in about 10% of the samples.
It’s RNA with only 1000 nucleotides.
Viroid-like colonists of human microbiomes Ivan N. Zheludev, Robert C. Edgar, Maria Jose Lopez-Galiano, Marcos de la Peña, Artem Babaian, Ami S. Bhatt, Andrew Z. Fire bioRxiv 2024.01.20.576352; doi: https://doi.org/10.1101/2024.01.20.576352
21.13 Diversity and Immunity
21.14 Statistically Modeling for Health Status
Zhu, J., Xie, H., Yang, Z. et al. Statistical modeling of gut microbiota for personalized health status monitoring. Microbiome 11, 184 (2023). https://doi.org/10.1186/s40168-023-01614-x
Statistical modeling of gut microbiota for personalized health status monitoring
We systematically developed a statistical monitoring diagram for personalized health status prediction and analysis. Our framework comprises three elements: (1) a statistical monitoring model was established, the health index was constructed, and the health boundary was defined; (2) healthy patterns were identified among healthy people and analyzed using contrast learning; (3) the contribution of each bacterium to the health index of the diseased population was analyzed. Furthermore, we investigated disease proximity using the contribution spectrum and discovered multiple multi-disease-related targets.
via Ken Lassessen
21.15 Eye Disease and Gut Microbiome
Some inherited eye diseases, including blindness may be caused by gut bacteria that
RB1 gene is key to controlling the integrity of the lower gastrointestinal tract, the first ever such observation. There, it combats pathogens and harmful bacteria by regulating what passes between the contents of the gut and the rest of the body.
The team found that when the gene has a particular mutation, dampening its expression (reducing its effect), these barriers in both the retina and the gut can be breached, enabling bacteria in the gut to move through the body and into the eye, leading to lesions in the retina that cause sight loss.
The research was conducted on mice by Professor Richard Lee (UCL Institute of Ophthalmology and Moorfields Eye Hospital NHS Foundation Trust).
https://www.cell.com/cell/abstract/S0092-8674(24)00108-9
21.16 Skin Microbiome and Attractiveness to Mosquitoes
Skin microbiome alters attractiveness to Anopheles mosquitoes
Staphylococcus 2 ASVs are four times as abundant in the highly-attractive compared to poorly-attractive group.
via Axios
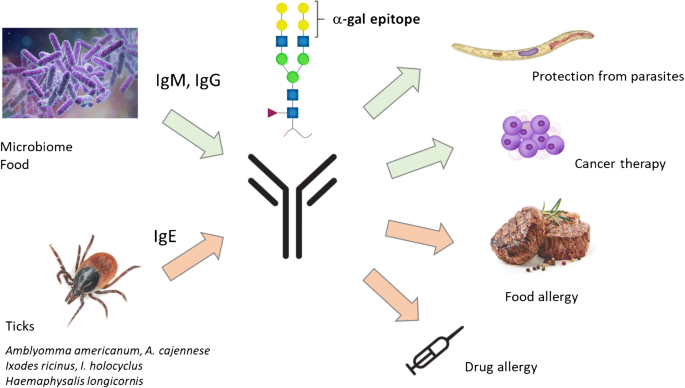
21.17 Ticks and Alpha-Gal
see Ticks, Alpha-Gal, Neu5gc and more
The CDC warns about
Alpha-gal syndrome (AGS) is a serious, potentially life-threatening allergic condition. AGS is also called alpha-gal allergy, red meat allergy, or tick bite meat allergy.
does it have an association with microbes?
21.18 Gut-Brain
a new study reveals the gut has a much more direct connection to the brain through a neural circuit that allows it to transmit signals in mere seconds.
21.19 Paleo Humans
Natural products from reconstructed bacterial genomes of the Middle and Upper Paleolithic from a German team that > Now, a new study from an interdisciplinary team has taken important steps to understanding stone age bacteria by sequencing genomes recovered from ancient dental calculus. The hardened tartar preserved bacterial fragments on the teeth of 12 Neanderthals and 34 humans that had lived anywhere from 102,000 to 150 years ago. Formed from plaque, this calculus fossilized during these humans’ lifetime, trapping genetic fragments inside.
Discover Magazine on Bacterial DNA from ancient humans
21.20 Microbiome uniqueness
See https://pubmed.ncbi.nlm.nih.gov/30150716/
A Chinese study that found microbiome patterns that predict health in one province don’t work in another.
He et al. (2018)
He Y, Wu W, Zheng HM, Li P, McDonald D, Sheng HF, Chen MX, Chen ZH, Ji GY, Zheng ZD, Mujagond P, Chen XJ, Rong ZH, Chen P, Lyu LY, Wang X, Wu CB, Yu N, Xu YJ, Yin J, Raes J, Knight R, Ma WJ, Zhou HW. Regional variation limits applications of healthy gut microbiome reference ranges and disease models. Nat Med. 2018 Oct;24(10):1532-1535. doi: 10.1038/s41591-018-0164-x. Epub 2018 Aug 27. Erratum in: Nat Med. 2018 Sep 24;: PMID: 30150716.
https://www.nature.com/articles/s42255-021-00348-0
see evernote
Wilmanski et al. (2021)
21.21 Methods
Greengenes2 unifies microbial data in a single reference tree
Studies using 16S rRNA and shotgun metagenomics typically yield different results, usually attributed to PCR amplification biases. We introduce Greengenes2, a reference tree that unifies genomic and 16S rRNA databases in a consistent, integrated resource. By inserting sequences into a whole-genome phylogeny, we show that 16S rRNA and shotgun metagenomic data generated from the same samples agree in principal coordinates space, taxonomy and phenotype effect size when analyzed with the same tree.
21.22 Mapping the Capacity of a Single Subject’s Microbiome to Metabolize Hundreds of Drugs
57 drugs that are transformed by the microbiome, with lots of variance from person to person.
https://www.cell.com/cell/fulltext/S0092-8674(20)30563-8#secsectitle0035
Javdan et al. (2020)
see ‘Most Life on Earth Is Dormant, After Pulling an ‘Emergency Brake’’ in https://www.quantamagazine.org/most-life-on-earth-is-dormant-after-pulling-an-emergency-brake-20240605↩︎
21.11.1 Social Transmission of Microbiomes
A 2024 study by Yale Network Scientist Nicholas Christakis says Your friends shape your microbiome — and so do their friends. They mapped the social networks and microbiomes of almost 2000 people in rural Honduras villages and found—sure enough—that people who interact regularly have more similar microbes than those who don’t.
Full text: Beghini, F., Pullman, J., Alexander, M. et al. Gut microbiome strain-sharing within isolated village social networks. Nature (2024). https://doi.org/10.1038/s41586-024-08222-1